Maize Projects
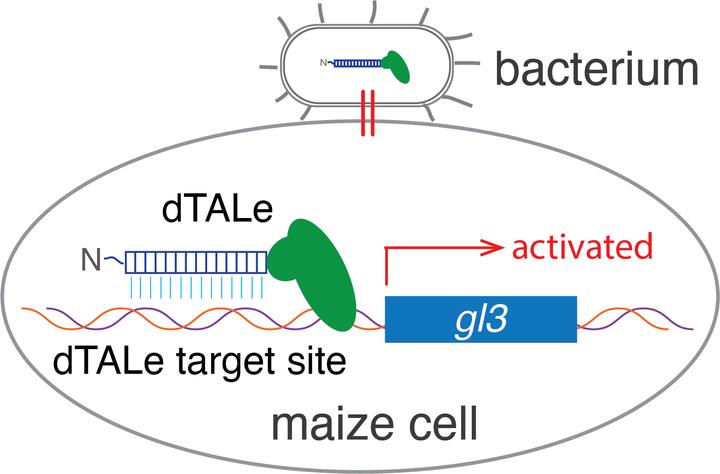 designer TALe
designer TALe
Maize Embryogenesis and Regeneration
CRISPRs provide great potential for the elucidation of maize gene functions and trait improvements. However, current genetic and genomic resources as well as transformation capacity in maize are insufficient for the full utilization of genome engineering tools. We are generating public resources that include a high-quality genome assembly of a highly transformation amenable maize inbred line A188 and double haploid (DH) lines from a cross of A188 with B73, a transformation recalcitrant inbred variety and the maize reference genome. We are genetically mapping the genetic elements controlling embryogenesis and regeneration. Our goal is to identify genes through map-based cloning with the assistance from multilayered gene regulatory networks, and apply the genes to enhance maize transformation.
Maize Diseases - Goss’s Wilts
Goss’s wilt is a maize disease, caused by Clavibacter michiganensis subsp. nebraskensis (CMN) that was first identified in 1969. The disease has not been a serious problem until recent re-emergence in the Great Plains region. While a number of Clavibacter-related diseases have been characterized, few genetic resources have been identified specifically for Goss’s wilt. We performed traditional GWAS, traditional QTL, XP-GWAS, XP-CNV to dissect disease resistance. Meanwhile, we performed genome sequencing for multiple Cmn strains. Our project goal is to provide extensive genetic information into plant resistance and bacterial virulence with regard to the re-emergence of Goss’s Wilt disease of maize. Our ultimate goal is to identify resistance genes in plant host and virulence elements in bacteria.
Genomic Structural Variation
Genomes contain structural variations (SV) that are comprised of unbalanced forms (copy number variation) and balanced forms (sequences in same copy number but at different genomic locations), and SV has been implicated in phenotype variation. However, SV often occurs on a scale that is difficult to be characterized with a single approach. We are attempting to combine multiple approaches, next-generation sequencing, genetic approach, and BioNano genome mapping technology to characterize SV.
Change in Gene Expression in Maize Under Abiotic Conditions
The genome-wide messenger RNA expression (mRNA), including splicing events, and small RNA (sRNA) expression are tractable at the unprecedented level with the utility of the next-generation sequencing technology. This project is designed to measure mRNA and sRNA from plant individuals under different controlled abiotic stresses. Through quantifying the change in the expression of genes, splicing sites, and sRNAs in response to abiotic conditions, we hope to find some candidates as the targets that can be used to enhance plant tolerance to extremely environmental conditions. sRNA and mRNA co-expression networks were also constructed to understand the change in expression correlation under drought condition.